I was looking at my site’s traffic, and I notice that an awful lot of people find my website by googling questions about the DNA shared between siblings.
I thought it might be useful to answer some of these questions and clear a few things up.
“What percent of my DNA is the same as my sister’s?”
“How much of my DNA is shared with a sibling?”
“How much DNA should brothers and sisters share?"
“How much of my DNA is shared with a sibling?”
“How much DNA should brothers and sisters share?"
The short answer is that siblings share an average of 50% of their DNA.
Here’s why siblings share 50% of their DNA.
Your mom has two copies of every piece of DNA and your dad also has two copies. To make a child, each parent passes on one copy of their DNA – consequentially, the child will also have two copies of that piece of DNA.
I have drawn an example below.
Let’s say that on this particular piece of DNA, you are outcome #3 – ‘green and purple’. You got a ‘green’ copy of DNA from your mom and a ‘purple’ copy from your dad.
Your parents can produce a child with any one of the four combinations that I drew. Take a look at how the four possible outcomes compare to your outcome (green and purple).
There is a one out of four chance (25%) that a new child of your parents would have an identical match as you (that is to say, they would also get a green gene from mom and a purple gene from dad).
There is a two out of four chance that the new child would be only half-identical to you. If the child was outcome #1, they would have an identical gene from your dad (purple) but a different gene from your mom (red instead of green like you). Outcome #4 is the same except reversed. Your sibling would have an identical gene from your mom (green) but a different gene from your dad (blue instead of purple).
There is a one out of four chance that the sibling would inherit a different copy of both genes. This is what happens with outcome #2. This sibling got a red copy of your mom’s DNA, while you got a green copy. He also got a blue copy of your dad’s DNA, while you got a purple copy instead.
To summarize, at any given point in the genome, there is one way to be fully identical with your sibling, two ways to be half-identical, and one way to be non-identical.
Said another way, across the entire genome, you will share about ¼ of your DNA fully identical with your sibling. You will share about ½ of your DNA half-identical with your sibling, and about ¼ of your DNA will be non-identical compared to your sibling.
25% identical DNA (100% match – same from mom AND dad) = .25 * 100 = 25%
50% half-identical DNA (50% match – same from mom OR dad but not both) = .50 * 50 = 25%
25% not identical DNA (0% match – different from both mom and dad) = .25 * 0 = 0%
25% + 25% + 0% = 50%. Siblings share an average of 50% of their DNA.
Okay, I’ll rephrase this: “Why do I only share 45% with my sister?”
50% is only an average percentage. This percentage can and does vary quite a bit. On the thread that I started on 23andMe about shared DNA percentages with relatives (23andMe members can read that thread here) the highest reported sibling percentage was 58.27% and the lowest was 40.99%. Sharing 45% with a sibling is lower than average, but it's nothing unusual.
"Will my siblings get the same 23andMe results?"

"Will my siblings get the same 23andMe results?"
Not really.
Some aspects of your 23andMe results will be the same – for example, your maternal and paternal haplogroups.
Most of your results will not be the same as your sibling’s results, because, like we discussed earlier, you and your sibling each get a unique combination of DNA from your parents. Your health results will most likely be somewhat similar, but they will not be identical. Same with Relative Finder – your results will be similar. Because you share ~50% of your DNA with a sibling, you will share roughly half of your relatives in common with your sibling (but around half will be different for each person).
Sort of. It depends on what you mean by 'closer DNA match'. Every child gets a complement of 23 chromosomes from each parent -- this does not vary. However, a father passes along a Y sex chromosome to a son, and an X sex chromosome to a daughter. In terms of size, the X chromosome (left) is a lot bigger than the Y chromosome (right). You could say, based on this size difference, that a father gives more DNA to his daughter compared to his son, and that therefore a daughter has a closer match to her father than her brother does. Some companies, such as 23andMe, will report that a man shares ~47% of DNA with his son and ~50% with his daughter. So in this respect, you could say that one sibling has a closer DNA match to the parent than the other sibling does.
However, something to keep in mind is that the parent-child relationship does not vary like other relationships do. For example, we already talked about the wide range of percentages that siblings can share (on my thread, they share anywhere from about 41% to 58%). Parents and children do not vary like this -- it's always 50% from mom, 50% from dad.
"Can DNA test if you are full sisters without the parents?""Can sisters determine if they are true sisters through DNA testing, even if both parents are dead?"
Yes, definitely.
Sibling relationships are very unambiguous, with or without the parents.
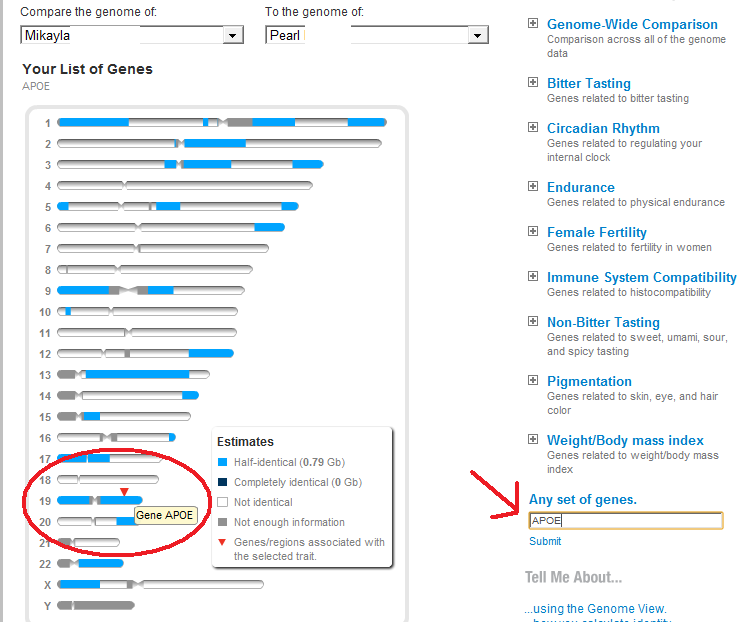
On 23andMe, sibling comparisons look like this. The percentage of DNA shared is approximately 50% and there are both fully-identical segments and half-identical segments (as well as some non-identical segments). No parents are needed to identify a sibling relationship on 23andMe.
For what it's worth, here's what a half-sibling comparison looks like.

For the most part, half-siblings share only half-identical segments -- no fully-identical segments. (An exception to this rule would be seen with maternal half-brothers on the X chromosome. Men have only one X chromosome, so any match between two men on the X would show as fully-identical.)

For the most part, half-siblings share only half-identical segments -- no fully-identical segments. (An exception to this rule would be seen with maternal half-brothers on the X chromosome. Men have only one X chromosome, so any match between two men on the X would show as fully-identical.)
Also, here's what a comparison between two unrelated people looks like.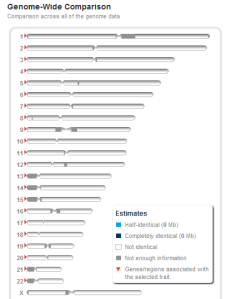
It's blank, with no shared segments.

It's blank, with no shared segments.
You can see that these various outcomes are very distinctive from one another. 23andMe can definitively determine whether two people are full siblings or not.